
Info
A.H.C. van Kampen
Work address
Bioinformatics Laboratory
Epidemiology & Data Science (EDS)
Amsterdam University Medical Centers (location AMC)
Meibergdreef 9, 1105 AZ Amsterdam UMC
Room J1B (J-building, floor 1B)
Biosystems Data Analysis
University of Amsterdam
Science Park 904, 1098 XH Amsterdam
Room C2-206
Websites:
https://bioinformaticslaboratory.eu/
https://github.com/EDS-Bioinformatics-Laboratory/ENCORE/wiki
Professional Career
After studying Chemistry and Informatics/chemometrics, I did my PhD at the Radboud University (1992-1996) on “The applicability of genetic algorithms to complex optimization problems in chemistry“. Subsequently, I moved to the Amsterdam UMC where I got involved in bioinformatics. Currently, he is the group leader of the Bioinformatics Laboratory, which is part of the department of Epidemiology and Data Science. I hold the chair Medical Bioinformatics at the Faculty of Medicine (Amsterdam University Medical Centers). I collaborate with the Biosystems Data Analysis group (UvA) where I am hosted for one day per week. I was scientific director of the Netherlands Bioinformatics Center (NBIC) from 2006 – 2010. Currently, he is member of the Uva Data Science Center steering committee and the UvA ICT for Research committee.
Research
The past 10 years the Bioinformatics Laboratory increasingly focused on systems medicine including computational modelling and (spatial) single-cell bioinformatics with a large focus on immunology. For example, in the context of Chronic Lymphocytic Leukemia (CLL), we develop AI/Machine learning approaches to extract relevant features from organoid images, and link those to single-cell molecular readouts to gain detailed insight into organoid development and treatment effects on tumor architecture and immune-stromal interactions. We also have been working of (single cell) B-cell and T-cell repertoire sequencing bioinformatics[1] in the context of auto-immunity (e.g., rheumatoid arthritis[2], and infection[3]).
We recently started efforts for molecular network reconstruction from single cell data aiming to obtain causal networks that can be used as input for mathematical modelling. In the past we developed and used various computational modelling approaches including (i) Monte Carlo simulations to investigate the effect of somatic hypermutations on the number of N-glycosylation sites in B-cell receptors [4], (ii) Petri nets for experimental design in multi-organ metabolite elimination pathways [5], (iii) a multiscale model using ordinary differential equations (ODEs) and agent-based modelling (ABM) to model plasma cell differentiation in the germinal center [6], (iv) Gillespie stochastic simulation algorithm to model plasma cell differentiation (unpublished), and (v) dynamic flux balance analysis to estimate flux profiles in metabolic networks [7]. In addition, we develop methods for the analysis LC-MS lipidomics data.
 |
| Figure 2. Multiscale Agent-base/ODE model of the germinal center. |
Finally, we aim to improve transparency and reproducibility of computational research, which is often challenging despite established guidelines and best practices. Translating these guidelines into practical applications remains difficult. We developed ENCORE [8], an approach to enhance transparency and reproducibility by guiding researchers in how to structure and document computational projects.
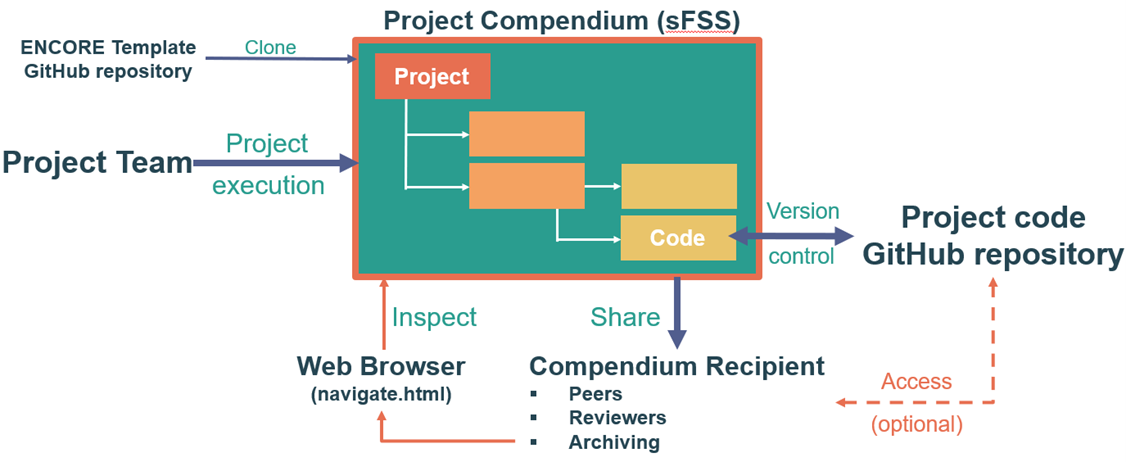 |
| Figure 3. ENCORE (ENhancing COmputational REsearch) setup. |
References
1. Balashova, D., et al., Systematic evaluation of B-cell clonal family inference approaches. BMC Immunol, 2024. 25(1): p. 13.
2. Pollastro, S., et al., Sensitive B-cell receptor repertoire analysis shows repopulation correlates with clinical response to rituximab in rheumatoid arthritis. Arthritis Res Ther, 2024. 26(1): p. 70.
3. Claireaux, M., et al., A public antibody class recognizes an S2 epitope exposed on open conformations of SARS-CoV-2 spike. Nat Commun, 2022. 13(1): p. 4539.
4. Vergroesen, R.D., et al., N-Glycosylation Site Analysis of Citrullinated Antigen-Specific B-Cell Receptors Indicates Alternative Selection Pathways During Autoreactive B-Cell Development. Front Immunol, 2019. 10: p. 2092.
5. Reshetova, P., et al., Using Petri nets for experimental design in a multi-organ elimination pathway. Comput Biol Med, 2015. 63: p. 19-27.
6. Merino Tejero, E., et al., Multiscale Modeling of Germinal Center Recapitulates the Temporal Transition From Memory B Cells to Plasma Cells Differentiation as Regulated by Antigen Affinity-Based Tfh Cell Help. Front Immunol, 2020. 11: p. 620716.
7. Willemsen, A.M., et al., MetDFBA: incorporating time-resolved metabolomics measurements into dynamic flux balance analysis. Mol Biosyst, 2015. 11(1): p. 137-45.
8. van Kampen, A.H.C., et al., ENCORE: a practical implementation to improve reproducibility and transparency of computational research. Nature Communications, 2024. 15(1): p. 8117.
Courses and topics
BSc Biomedische Wetenschappen
Academische Vaardigheden: Miniscripties.
Genetica en Evolutie Theorie: Highlight college.
OMICS in de biomedische wetenschappen: Mathematical Modelling.
MSc Computational Sciences
Bioinformatics Project I: Modelling and bioinformatics.
Bioinformatics Project II: Modelling and Bioinformatics.
MSc Biomedical Sciences
Biomedical Systems Biology: Models, Systems Biology.

