
Info
Jordan Earle
Work Address
Biosystems Data Analysis, University of Amsterdam
Science Park 904, Room C2-203, 1098 XH, Amsterdam
E-mail: j.a.earle@uva.nl
Professional Career
I started my career with a Mechanical Engineering BSc with a concentration in modeling of dynamic systems back in 2013. I worked for numerous engineering companies over a variety of field including solar, automotive, civil, and pharmaceutical.
In my last engineering position, I worked for a custom bone replacement implant company. I was brought on to create standardized templates, but while creating these, I found a lot of the work I and the other engineers were doing could be automated. I spent some time learning how to code, and quickly created a few automated templates for designs which became a staple for the company, saving the designers and CAD engineers hours of time per design.
As I created more templates and software for the company, my aspirations grew and I decided I wanted to have a formal education in Computer Science and AI. I came to the UvA in 2018 while working for my previous company in the UK to study Computational Science and Artificial Intelligence, so I could further automate the designs for my company. This plan quickly changed when I rediscovered my love for research, and I quickly changed my plan to go into academia. I graduated in 2022 with an MSc in Computational Science with a focus in AI from the University of Amsterdam.
For my MSc thesis I investigated the presence of a functional hierarchy of brain regions in macaque monkeys from Local Field Potential data using granger causality. This was done by looking at different regions and determining the interactions and changes in interactions between the regions during different behavioral states. I also used a multi-level model to simulate these interactions. This project cemented my interest in academia and after a brief stint in industry as a consultant I started working with the BDA group in 2024 with Crop XR to model the effects of (a)biotic stresses on the production of Glucosinolates in Arabidopsis thaliana.
Research
In my project I will be modeling the effects of (a)biotic stresses on the production of Aliphatic and Indolic Glucosinolates (GSLs) in Arabidopsis thaliana. I will be using a hybrid modeling approach, using both traditional mechanistic models (such as Michaelis Menten) and AI approaches to model the Glucosinolate concentrations. I will be attempting to identify and reproduce the important pathways in silico and show how these concentrations evolve over time depending on the mRNA levels produced by the changes in conditions.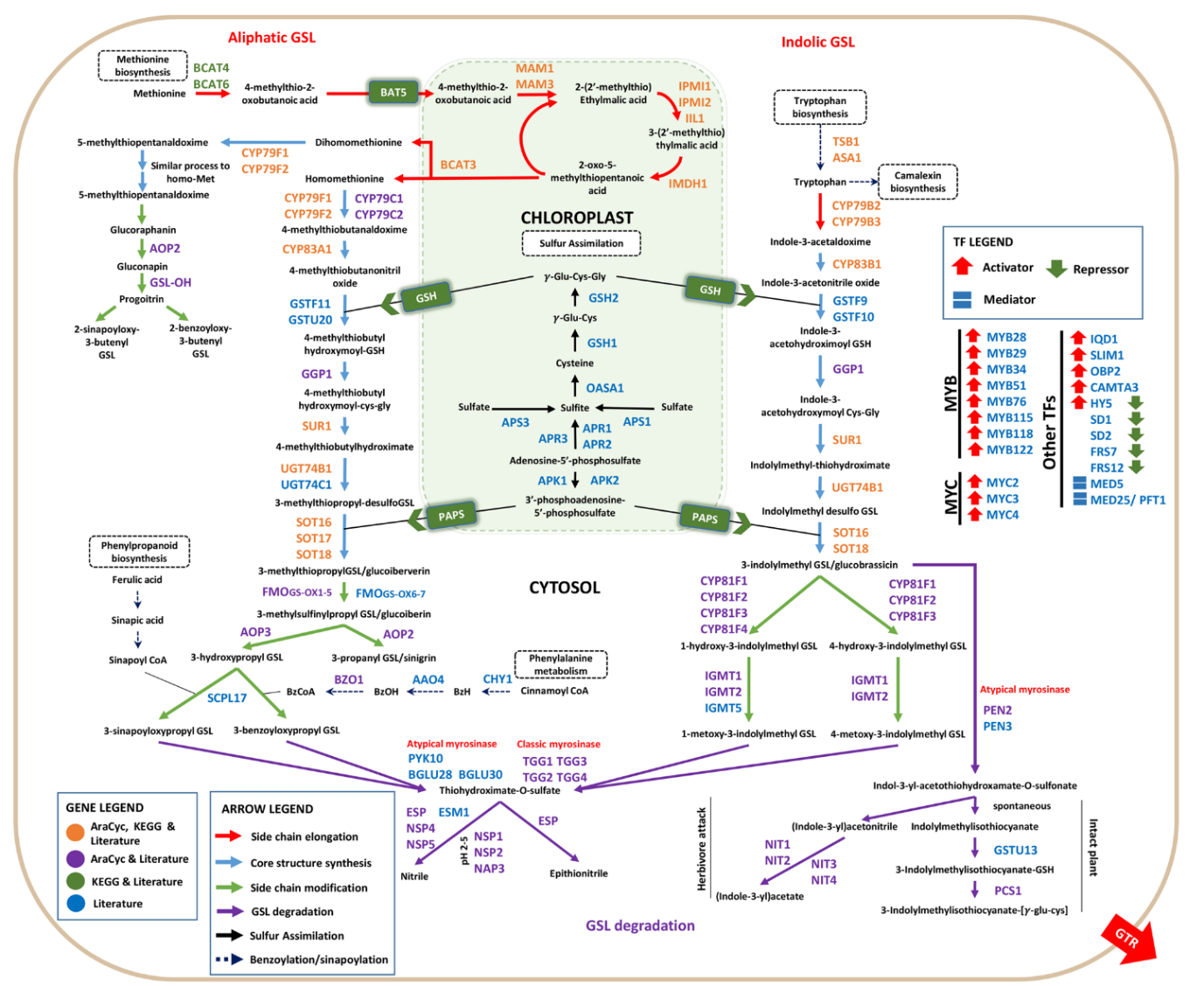
(Image from Harun et al. 2020)
