
Info
Aalt D.J. van Dijk
Work address:
Group Biosystems Data Analysis, University of Amsterdam
Science Park 904, Room C2-202, 1098 XH, Amsterdam
Email: a.d.j.vandijk@uva.nl
Phone +31 20 525 5519
Websites:
https://sils.uva.nl/profile/d/i/a.d.j.van-dijk/a.d.j.van-dijk.html
Professional Career
After studying Chemistry, I did my PhD at Utrecht University (2002-2006) on “Computational docking for the study of biomolecular interactions”. Subsequently, I worked for several years at Wageningen University and Research. Initially, I was positioned at the Bioscience department of Wageningen Research. Since 2016 I was part of the Bioinformatics group of Wageningen University, first as assistant professor and since 2021 as associate professor. In 2023 I started as professor “Data Analysis in the Life Sciences” at the Swammerdam Institute.
Research
My research involves development of machine learning approaches, and application of these methods in close collaboration with experimental colleagues. Large amounts of data are being produced in the life sciences, at various levels (e.g. genomics, phenotypic, ecological), and much of this data is quite unstructured. Machine learning is often applied as a black box, which can be sufficient if all one cares about is good prediction performance. However, it can be very fruitful to learn from the machine learning model itself. This allows for example to generate testable hypotheses, leading in turn to improved understanding of the system of study. Importantly, this also gives rise to opportunities to modify the system in order to obtain specific desired behaviour.
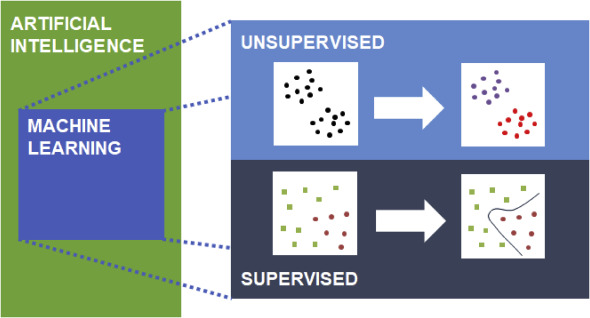 |
|
Figure 1, Machine learning involves learning predictive patterns |
Recent examples where I used machine learning in this way include PhD projects on the prediction of enzyme product specificity, the prediction of the pathogenic effect of protein coding variation, and the prediction of crossover occurrence in genomic locations for various plants. An important shared characteristic in these projects is that sufficient understanding of the application domain is essential to enable effective collaboration with domain scientists. Often, specific insights into the domain will be leading in how machine learning approaches can best be developed for specific purposes – standard solutions do not suffice.
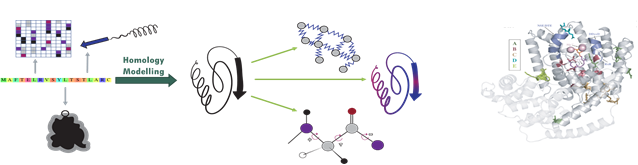 |
|
Figure 2, A recent example of machine learning methodology involves using protein structures as input to predict functions and interactions (left). Using interpretable machine learning allows to find regions in a protein which are important for specific functions (right). |
Courses and topics
BSc Biomedische Wetenschappen
Advanced Genomics Analysis: Omics data analysis for biomedical sciences.
MSc Brain and Cognitive Sciences
Cognitive Data Science: Genes, Brains and Behaviour: Omics data analysis for brain and cognitive sciences.
MSc Biomedical Sciences
Genomics: Machine learning on genomics data.
MSc Biological Sciences
Current Topics in Biology: Molecular networks and machine learning.
